Sansa Tarql
TARQL (https://tarql.github.io/) is a tool for mapping tabular data to RDF via SPARQL queries. This works by first converting each CSV row to a SPARQL binding and then evaluating the SPARQL query w.r.t. to that binding. Sansa features an Apache Spark based re-implementation that allows for processing a CSV file in parallel.
The implementation is also based on ApacheJena and our own “JenaX” extension project.
Important
Parallel processing requires specifying --out-file and/or --out-folder! If neither argument is given then output is streamed to STDOUT using a single thread (using RDD.toLocalIterator).
TARQL Compatibility
The design of Sansa tarql is aimed to ease the transition to or from the original tarql. Therefore, many options of the original are supported.
Notable extensions:
- The JenaX extension functions are available
Notable limitations:
- The extension functions
tarql:expandPrefix(?prefix)andtarql:expandPrefixedName(?qname)are not yet supported.
CLI Options
Usage: sansa tarql [-hHrstuVX] [--accumulate] [--header-row] [--iriasgiven]
[--ntriples] [--optimize-prefixes] [--out-overwrite]
[--pretty] [-d=<delimiter>] [-e=<encoding>] [-o=<outFormat>]
[--oup=<deferOutputForUsedPrefixes>] [--out-file=<outFile>]
[--out-folder=<outFolder>] [-p=<quoteEscapeChar>]
[-q=<quoteChar>] [--sort-partitions=<sortPartitions>]
[--unique-partitions=<uniquePartitions>]
[--header-naming=<columnNamingSchemes>]...
[--op=<outPrefixes>]... <inputFiles>...
Map one or more CSV files to RDF via a single SPARQL query
<inputFiles>... tarql query file following by one or more csv file
--accumulate Accumulate every row's intermediate mapping output
in a dataset which subsequent mappings can query
-d, --delimiter=<delimiter>
Delimiter
-e, --encoding=<encoding> Encoding (e.g. UTF-8, ISO-8859-1)
-h, --help Show this help message and exit.
-H, --no-header-row no header row; use variable names ?a, ?b, ...
--header-naming=<columnNamingSchemes>
Which column names to use. Allowed values: 'row',
'excel'. Numerics such as '0', '1' number with
that offset. If there are no header rows then
'row' is treated as 'excel'. Column names are
unqiue, first name takes precedence.
--header-row Input file's first row is a header with variable
names (default)
--iriasgiven Use an alternative IRI() implementation that is
non-validating but fast
--ntriples Tarql compatibility flag; turns any quad/triple
based output to nquads/ntriples
-o, --out-format=<outFormat>
Output format. Default: null
--op, --out-prefixes=<outPrefixes>
Prefix sources for output. Subject to used prefix
analysis. Default: rdf-prefixes/prefix.cc.
2019-12-17.ttl
--optimize-prefixes Only output used prefixes (requires additional
scan of the data)
--oup, --out-used-prefixes=<deferOutputForUsedPrefixes>
Only for streaming to STDOUT. Number of records by
which to defer RDF output for collecting used
prefixes. Negative value emits all known
prefixes. Default: 100
--out-file=<outFile> Output file; Merge of files created in out-folder
--out-folder=<outFolder>
Output folder
--out-overwrite Overwrite existing output files and/or folders
-p, --escapechar=<quoteEscapeChar>
Quote escape character
--pretty Enables --sort, --unique and --optimize-prefixes
-q, --quotechar=<quoteChar>
Quote character
-r, --reverse Sort ascending (does nothing if --sort is not
specified)
-s, --sort Sort data (component order is gspo)
--sort-partitions=<sortPartitions>
Number of partitions to use for the sort operation
-t, --tabs Separators are tabs; default: false
-u, --unique Make quads unique
--unique-partitions=<uniquePartitions>
Number of partitions to use for the unique
operation
-V, --version Print version information and exit.
-X Debug mode; enables full stack traces
Examples
The sansa command is available either by installing the debian/rpm packages or by running the jar bundle directly using java -jar sansa-version.jar
Depending on the Java version you may need to add --add-opens declarations as documented here.
- Basic Invocation
sansa tarql mapping.rq input.csv --out-file /tmp/output.ttl - Faster processing of the IRI() function by disabling validation. Only use this if you can ensure the generated IRIs will be valid.
sansa tarql mapping.rq input.csv --iriasgiven --out-file /tmp/output.ttl -
Add the
--out-overwriteoption to delete any prior hadoop output. This operation uses a safe-guard against accidental recursive folder deletion: If the output folder contains any non-hadoop files then an exception will be raised. - When using the deb/rpm packges then options for hadoop/spark can be supplied via the
JAVA_OPTSenvironment variable.JAVA_OPTS="-Dspark.master=local[4]" sansa tarql mapping.rq input.csv --out-overwrite --out-folder /tmp/out-folder - When running directly with
javathen the hadoop/spark options can be provided as usual via-D:java -Dspark.master=local[4] -jar sansa-version.jar tarql
Minimal Example with Docker
A docker image of sansa is available under aklakan/sansa.
docker pull aklakan/sansa
echo -e 'http://example.org/foo,foo\nhttp://example.org/bar,bar' > /tmp/data.csv
echo "CONSTRUCT { ?s <http://www.w3.org/2000/01/rdf-schema#label> ?b } { BIND (IRI(?a) AS ?s) }" > /tmp/mapping.rq
docker run -v /tmp:/data -it aklakan/sansa tarql --delimiter , --no-header-row /data/mapping.rq /data/data.csv --out-format ntriples
- Remark: The presence of
--delimiter ,is due to the univocity CSV parser incorrectly auto-detecting it as:.
Performance Comparison
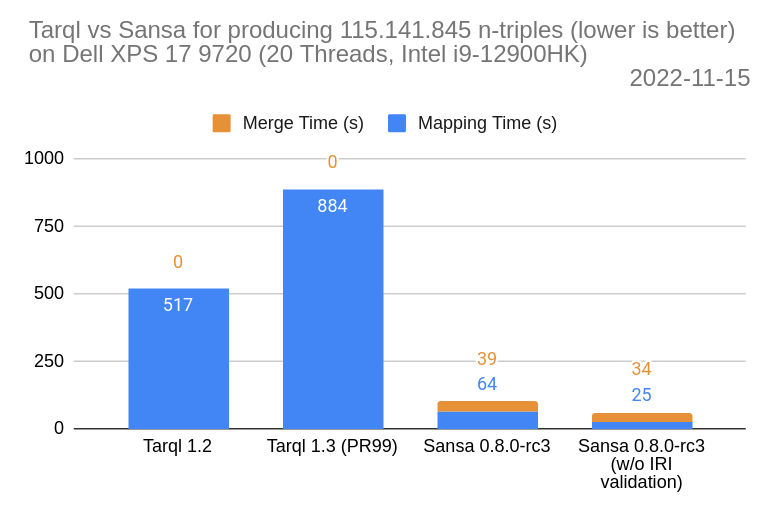
- Tarql 1.3 is based on tarql pull request 99 which uprades the code base to Jena 4.
- Mapping time is the time needed to write out the whole mapping once. In the hadoop/spark setting multiple files are written out at once.
- Merge time is the time needed to concatenate the prior output into a single file.
- Although a thorough analysis of time difference between tarql 1.2 (Jena 3) and tarql 1.3 (Jena 4) has yet to be conducted, it is most likely related to IRI processing.
- Jena 4’s IRI function not only adds validation overhead but also global synchronization which impacts performance of parallel processing (see JENA-1470).
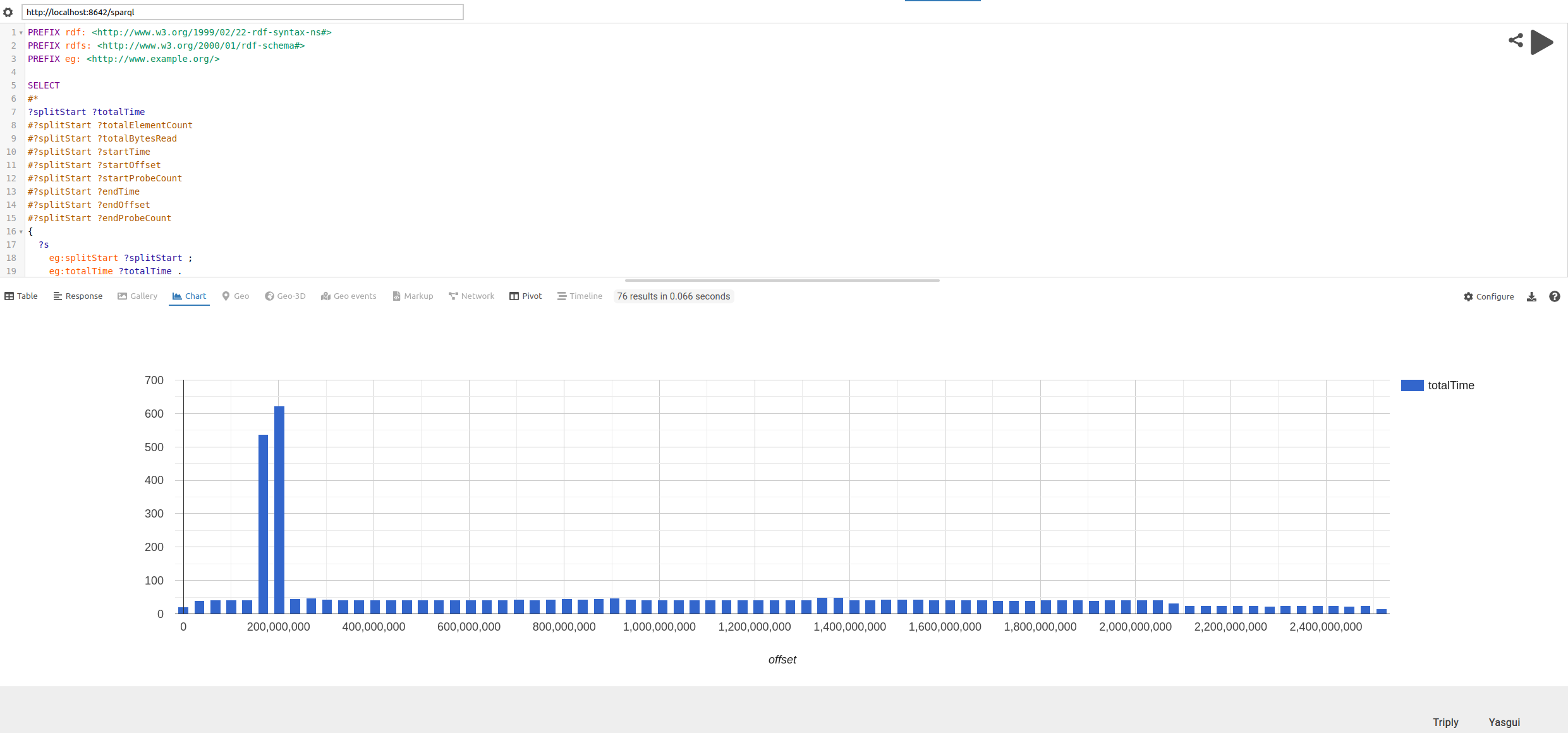
Inspecting CSV files
The CSV parser is capable of parsing multiline CSV files, however there may be issues in the data or bugs in the parser.
The sansa analyze csv command can be used to obtain per-split report in RDF for a given CSV file. This report includes information such as the number of records produced, the time it took and the number of
probing attempts to find a region offset.
sansa analyze csv --out-file stats.ttl input.csv
sansa anylze csv --out-file stats.ttl --delimiter , --encoding utf-8 input.csv
The output of the stats file can be processed with the following sparql query:
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX eg: <http://www.example.org/>
SELECT
*
#?splitStart ?totalTime
#?splitStart ?totalElementCount
#?splitStart ?totalBytesRead
#?splitStart ?startTime
#?splitStart ?startOffset
#?splitStart ?startProbeCount
#?splitStart ?endTime
#?splitStart ?endOffset
#?splitStart ?endProbeCount
{
?s
eg:splitStart ?splitStart ;
eg:totalTime ?totalTime .
OPTIONAL { ?s eg:totalBytesRead ?totalBytesRead }
OPTIONAL { ?s eg:totalElementCount ?totalElementCount }
OPTIONAL { ?s eg:regionStartProbeResult/eg:totalDuration ?startTime }
OPTIONAL { ?s eg:regionStartProbeResult/eg:candidatePos ?startOffset }
OPTIONAL { ?s eg:regionStartProbeResult/eg:probeCount ?startProbeCount }
OPTIONAL { ?s eg:regionEndProbeResult/eg:totalDuration ?endTime }
OPTIONAL { ?s eg:regionEndProbeResult/eg:candidatePos ?endOffset }
OPTIONAL { ?s eg:regionEndProbeResult/eg:probeCount ?endProbeCount }
}
ORDER BY ?splitStart
One way to visualize this data is as follows:
-
Start a local SPARQL endpoint using the triple store of your choice. For example, with the RDF Processing Toolkit the command
rpt integrate --server stats.ttlstarts an endpoint underhttp://localhost:8642/sparql. -
Visit Yasgui and configure it to your local endpoint
-
Copy&Paste the SPARQL query above. By default this should show the statistics as a table. By selecting the
charttab in Yasgui and adapting the projection of the query the different attributes can be visualized.
An example visualization of an issue where the parsing of two splits takes an excessive amount of time is shown below: